
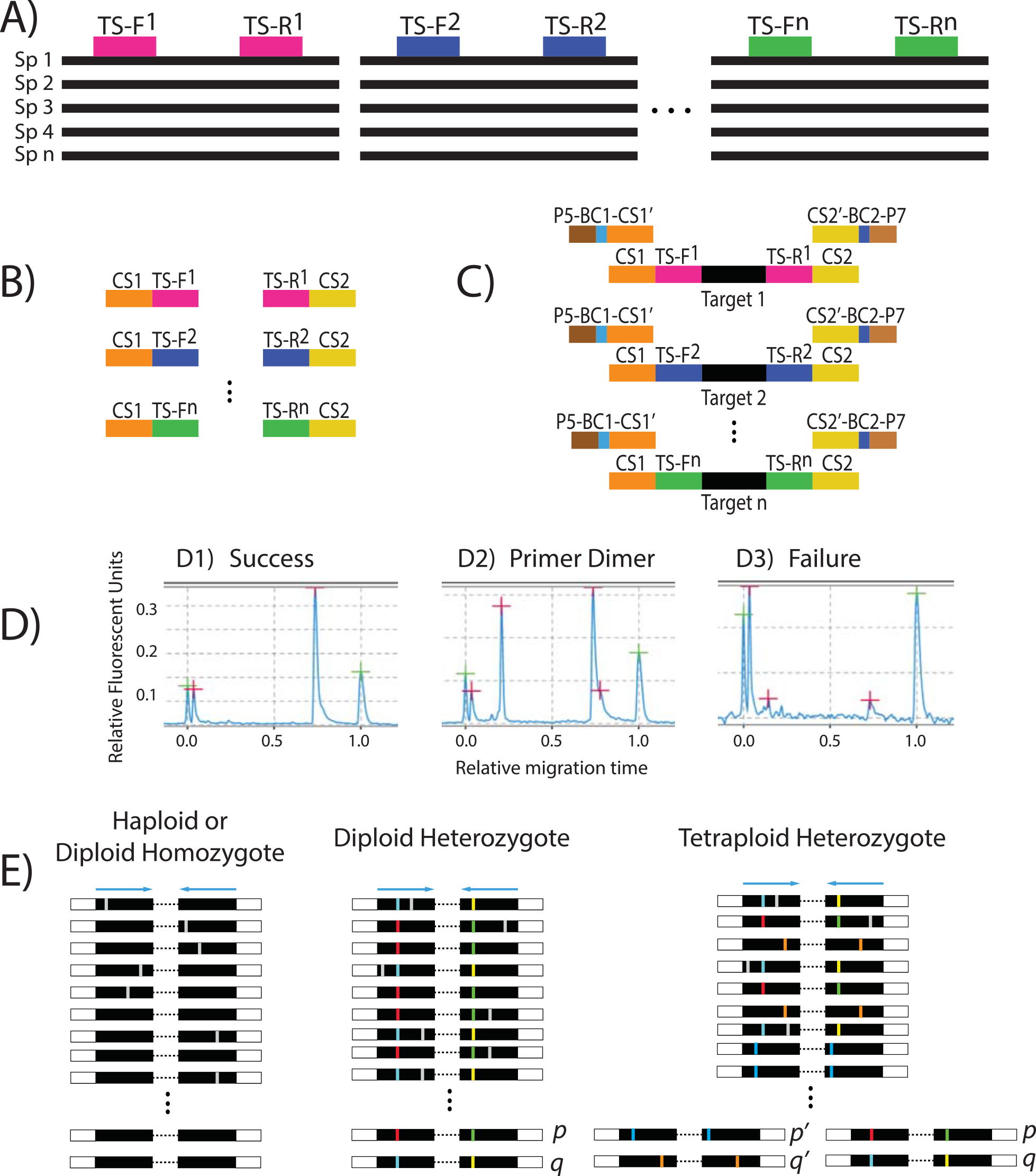
A new paper from my lab detailing a high-throughput approach for phylogenetic dataset collection using targeted, microfluidic PCR and Illumina sequencing came out this week in PLoS ONE. This was a collaboration between Simon Uribe-Convers and Matt Settles (former Director of the UI IBEST Genomics Core, now at UC Davis). We think that this approach has a lot of advantages over other genomic approaches for obtaining large, multigene datasets for phylogenetic analyses, especially at lower taxonomic scales. All of the code for analyzing the resulting data can be found on Matt’s GitHub site.
